This tutorial was originally presented as part of a 3-day intensive Data Science Workshop at UMass Amherst in June 2015. The materials for this workshop are all available on Github at the following link: https://github.com/matthewjdenny/ISSR_Data_Science_Summer_Summit_15
Overview
R can be perfectly fast for some things but sometimes too slow for other things. In general, R is not the fastest or most memory efficient language out there -- but it is very easy to use, to share, and makes very pretty output. C++ is very fast, but not so easy to use, and was not designed with making pretty output in mind. However, when we put the two together in a smart way, we can both go fast and write code that is relatively easy to use.
Requirements for using C++ code with R
If you are using a Mac, you will need to start by making sure you have Xcode + developer tools installed or you will not be able to compile the C++ code that is used in the samplers for this package. You will need to go here: https://developer.apple.com/xcode/downloads/ and then select the link to the additional downloads page which will prompt you to enter you apple ID. This will let you download the developer tools. This requirement is not unique to any particular package, but is necessary for all packages that use Rcpp.
If you are using certain version of OSX (at least Mavericks, but possibly others), you will also likely get an -lgfortran or -lquadmath error the first time you try to compile some C++ code using R. This error is detailed here, along with very nice instructions for fixing it. The simple solution is to open up a Terminal (by going to Applications -> Utilities -> Terminal) and paste the following two lines of code into the terminal. Note that by using the sudo command, you will be asked to enter your password. If you encounter an error, you can check out this page for instructions on enabling sudo on a mac. Once you have entered the code (if you did not get any error messages) then you should be fine to try sourcing C++ code again.
curl -O http://r.research.att.com/libs/gfortran-4.8.2-darwin13.tar.bz2
sudo tar fvxz gfortran-4.8.2-darwin13.tar.bz2 -C /
If you are using a Windows machine, you will need to make sure you have the latest release of R (3.2.0+) and will also need to install the Rtools library (available here http://cran.r-project.org/bin/windows/Rtools/) before you can use any packages with C++ code in them. Note that you should choose whichever is the latest version of Rtools when asked to select a version on the website. I have found that some people encounter strange errors where R keeps asking them to install Rtools repeatedly. If this is the case for you, I suggest you try restarting your computer as this tends to be a problem with certain older versions of Windows and not a general problem for Windows (it works just fine for the vast majority of folks). It is also highly advised that you use RStudio to download and install the package as it seems to play nicer with Rcpp under Windows. You may also want to visit this blog post which has more information on making C++ work with R under Windows if you end up encountering any weirdness.
If you are using a Linux distro, make sure you have a C++ complier installed, but in general, you should not run into as many issues
Rcpp, RcppArmadillo and RStudio
This is where Rcpp comes in. It is an R package that makes incorporating C++ code with R code pretty straightforward. The good folks over at RStudio have also baked in a lot of support for Rcpp right into RStudio, and have included a pretty useful debugger to boot. RcppArmadillo is an add-on package that gives you access to tons of useful linear algebra functionality in C++. In particular it makes passing in and working with arrays, matrices and vectors pretty easy. Here are some resources to get you started:
- The official website for Rcpp is -- http://www.rcpp.org/
- Dirk Edelbuettel has a great site for all things R check out the code and blog sections. He is the creator of the Rcpp package among many others. Here is the link: http://dirk.eddelbuettel.com/
- Hadley Wickham's Advanced R book has a nice chapter on Rcpp available here: http://adv-r.had.co.nz/Rcpp.html
- You can check out the Armadillo docs here: http://arma.sourceforge.net/docs.html. This is where I go to look up functions to see how to use them or if they exist.
Getting started
To get started writing C++ code using Rcpp, you will definitely want to check out the short tutorial at this link, which provides some nice examples of how to write a C++ function and debug it using RStudio. The basic idea can be illustrated in two screenshots. We start by creating a new file in RStudio by clicking on the doc+ icon in the top left corner of the screen and select C++ File from the drop down menu.
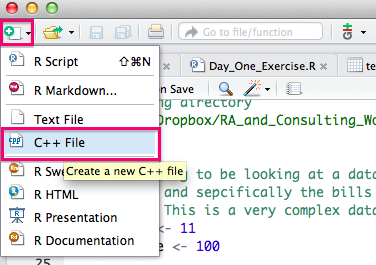
A sample timesTwo* C++ program will pop up. You will now need to save that file somewhere. Once you have done so, you can click on the Source button and you will see that RStudio automatically calls Rcpp::sourceCpp() on the file.
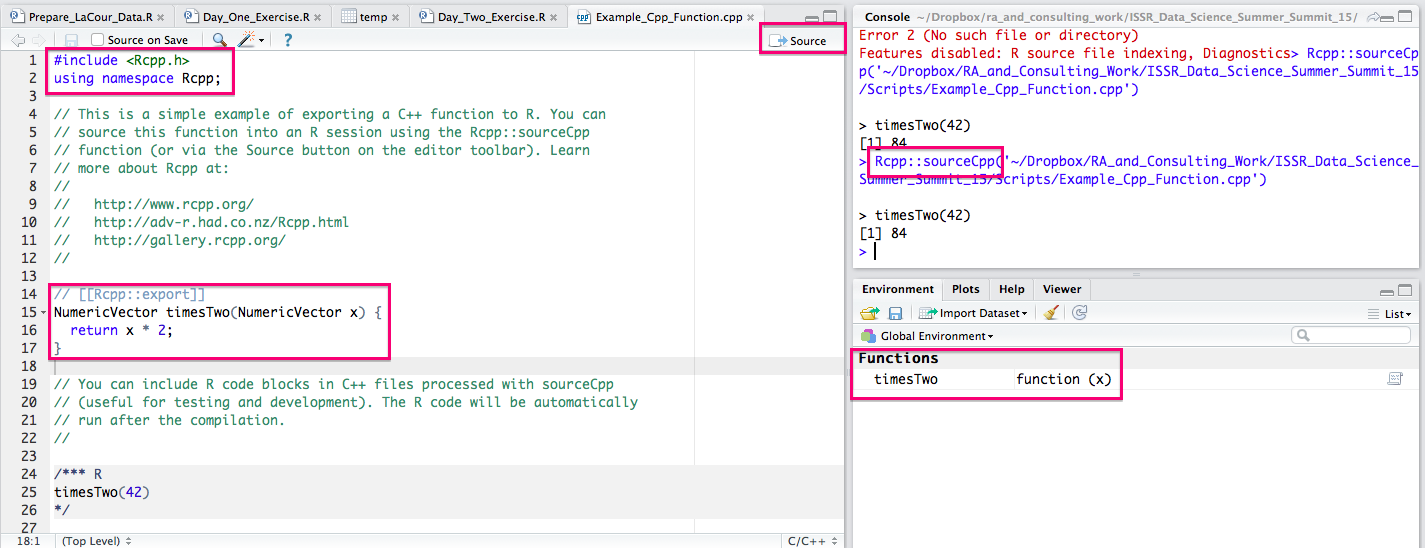
This will compile the function and make it so we can access the C++ code from R -- super easy and handy! This is all we need to get started. You will inevitably encounter errors and probably want to distribute your code, but for now, you can simply have access to blazing fast C++ functions on your own computer. Here is your new function:
#include <Rcpp.h>
using namespace Rcpp;
// This is a simple example of exporting a C++ function to R. You can
// source this function into an R session using the Rcpp::sourceCpp
// function (or via the Source button on the editor toolbar). Learn
// more about Rcpp at:
//
// http://www.rcpp.org/
// http://adv-r.had.co.nz/Rcpp.html
// http://gallery.rcpp.org/
//
// [[Rcpp::export]]
NumericVector timesTwo(NumericVector x) {
return x * 2;
}
// You can include R code blocks in C++ files processed with sourceCpp
// (useful for testing and development). The R code will be automatically
// run after the compilation.
//
/*** R
timesTwo(42)
*/
The Armadillo data structures + a template function
The Armadillo data structures provided by the RcppArmadillo package are really nice and easy to use, but they are also smart to use right off the bat. The base Rcpp package provides its own data structures that can be passed easily between R and C++, but so can the Armadillo data structures. Additionally, the Armadillo data structures are native C++ data structures while the Rcpp data structures are "shallow". What this means is that if you are doing tons of writing to really large Rcpp data structures (say writing lots of big matrices to an Rcpp::List object of length 100,000) you can actually run into some serious and very difficult to diagnose segfaults and other bad errors. I ran into a number of very hard to debug problems with by Rcpp code failing after running for several hours while working on the first version of a big machine learning algorithm a few years back, but after switching to Armadillo data structures, these issue went away. Lets take a look at some of the dat structures you can pass in. What follows is essentially the template I use for pretty much all of my Rcpp functions.
#include <RcppArmadillo.h>
//[[Rcpp::depends(RcppArmadillo)]]
using namespace Rcpp;
//[[Rcpp::export]]
List My_Function(
double my_double,
int some_number,
arma::vec a_vector,
arma::mat example_matrix,
arma::cube my_array,
List some_vectors
){
double new_double = 100*my_double;
List to_return(1);
to_return[0] = new_double;
return to_return;
}
Note that you no longer use the:
#include <Rcpp.h>
statement as in the example code provided by RStudio, but replace it with:
#include <RcppArmadillo.h>
//[[Rcpp::depends(RcppArmadillo)]]
Some other things to note:
This statement tells us that we can call any Rcpp constructs by their given name without the
Rcpp::prefix:using namespace Rcpp;We also put one of these statements before each function we want to make available to R:
//[[Rcpp::export]]We can also define multiple C++ functions in the same file (not necessarily recommended unless some of them will be used by the main function), so we can put one in front of each one we want to make visible.
Now lets take a look at the different objects we can pass in. Note that for all of them, we have to specify their type as we define the argument. This is a feature of C++ that is different from R where we just create objects without having to specify their type.
For decimal numbers like
1.2347we need to use thedoubledeclaration, followed by the name of the argument (e.g..my_double)For integers (whole numbers) like
26we use theintdeclaration, followed by the argument.For numeric vectors, we use the
arma::vecdeclaration, followed by the argument. This code should crash if you try to pass in anything other than a numeric vector (can contain doubles or integers).For numeric matrices, we use the
arma::matdeclaration, followed by the argument. Again, make sure it is just numbers in there.Things used to be a bit trickier for arrays (3d or greater). What gets constructed in c++ is an
arma::cube, but we had to pass in aNumericVectorobject which then got turned into anarma::cubeinternally using the following block of code. This seems a bit odd but it works if you are using an older version of Rcpp. However, since around 2016, it has been possible to simply pass in anarma::cubedirectly. However, if you ever run into a problem, this code can be a nice workaround:IntegerVector dim = array.attr("dim"); arma::cube my_array(array.begin(),dim[0], dim[1], dim[2], false);Finally, we can pass in lists of numbers, vectors or matrices (or arrays I suppose) using the
Listdata type. This is the only Rcpp data structure I use -- as Armadillo does not provide one, and in general as long as we do not try to do too much with it, we should be ok.I also tend to specify the return value as a
Listso that we can stick whatever values we want in it.List My_Function()This works well and again if you keep it shorter than say 1,000 entries for what you return you should not hit any snags with weird memory stuff.
One other important point with C++ is that how you initialize a double/int/vector/matrix/array matters. Under the hood, C++ will just go and grab some spaces in your RAM, and if those spaces have non-zero numbers in them, so will the entries in you vector (for example). This will not matter if you overwrite all of the values right off of the bat, but if you are constructing a distribution or something like that, this can spell trouble! Fortunately Armadillo has the arma::zeros to come to the rescue.
arma::vec new_vec = arma::zeros(len);
arma::mat new_mat = arma::zeros(len,len);
arma::cube new_array = arma::zeros(len,len,len);
One other thing you should note if you have not already is that every line of C++ code must be terminated with a ; or the code will not compile. Its just one of those things.
Some Examples
Lets take a look at a couple of example Rcpp functions I have written for different applications to start getting the hang of looping and other related concepts.Here is a function that I wrote about in a blog post here, which calculates the mutual information of an arbitrary joint distribution.
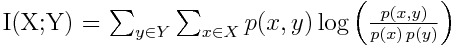
You can read more about mutual information in that post, or by checking out the wikipedia page, but what is important is that we have to traverse all the entries of a matrix and calculate some quantity. Note that we will need to use loops, which are defined as follows in C++ :
for(int i = 0; i < N; ++i){
//do some stuff
}
Lets take a look at the function:
#include <RcppArmadillo.h>
#include <cmath.h>
//[[Rcpp::depends(RcppArmadillo)]]
using namespace Rcpp;
// [[Rcpp::export]]
double Mutual_Information(
arma::mat joint_dist
){
joint_dist = joint_dist/sum(sum(joint_dist));
double mutual_information = 0;
int num_rows = joint_dist.n_rows;
int num_cols = joint_dist.n_cols;
arma::mat colsums = sum(joint_dist,0);
arma::mat rowsums = sum(joint_dist,1);
for(int i = 0; i < num_rows; ++i){
for(int j = 0; j < num_cols; ++j){
double temp = log((joint_dist(i,j)/(colsums[j]*rowsums[i])));
if(!std::isfinite(temp)){
temp = 0;
}
mutual_information += joint_dist(i,j) * temp;
}
}
return mutual_information;
}
Another thing to note here is that function returns do not get enclosed in () like they do in R. We can try compiling it and you will see it pop up in the Functions pane of RStudio. Now lets take a look at another function I wrote that finds the unique words in a corpus of documents and counts the number of times each unique word appears.
#include <RcppArmadillo.h>
#include <string>
//[[Rcpp::depends(RcppArmadillo)]]
using namespace Rcpp;
// [[Rcpp::export]]
List Count_Words(
int number_of_documents,
List Document_Words,
arma::vec Document_Lengths
){
List to_return(3);
int total_unique_words = 0;
arma::vec unique_word_counts = arma::zeros(250000);
std::vector<std::string> unique_words(250000);
for(int n = 0; n < number_of_documents; ++n){
Rcpp::Rcout << "Current Document: " << n << std::endl;
int length = Document_Lengths[n];
std::vector<std::string> current = Document_Words[n];
for(int i = 0; i < length; ++i){
int already = 0;
int counter = 0;
while(already == 0){
if(counter == total_unique_words){
unique_words[counter] = current[i];
unique_word_counts[counter] += 1;
total_unique_words += 1;
already = 1;
}else{
if(unique_words[counter] == current[i]){
unique_word_counts[counter] += 1;
already = 1;
}
}
counter +=1;
}
}
}
to_return[0] = total_unique_words;
to_return[1] = unique_words;
to_return[2] = unique_word_counts;
return to_return;
}
There are a couple of new things in this function. First, we are printing stuff to the R console using Rcpp::Rcout, a newly introduced function in Rcpp that make printing seamless and easy. We simply have to use << to separate the things we want to print (which can include variables), and then use an end line statement at the end std::endl. Here is some example code:
Rcpp::Rcout << "Variable Value: " << my_variable << std::endl;
You can find out more information on Rcout by checking out this tutorial. The other new thing this piece of code introduces is the vector of strings, which can be specified by using the std::vector<std::string> declaration followed by the name of the string vector you want to create. We need to use the standard vector class because as far as I know, the Armadillo vectors do not support strings. One of the cool things we can do is pass in an Rcpp::List object full of string vectors and then assign each of these vectors to a std::vector<std::string> C++ object and everything just works. We can also stick these back in a Rcpp::List and then return them to R without any trouble. Here is the example block of code:
std::vector<std::string> my_string_vector(1000);
Defining Sub-Functions
While writing a bunch of nested loops is great and all, we may also want to be able to write C++ function that can be used by other C++ functions just like we would with R code. To do this, you will need to define your own namespace before you can define sub-functions to be used by your other functions. This namespace will give a prefix to any functions you define so C++ can know what you are talking about. In the example below, the cdf() function would get called in my Rcpp program by mjd::cdf(). These are three functions that calculate the erf, pdf, and cdf of draws from a normal distribution.
#include <RcppArmadillo.h>
#include <cmath.h>
//[[Rcpp::depends(RcppArmadillo)]]
using namespace Rcpp;
namespace mjd {
// Returns the erf() of a value (not super precise, but ok)
double erf(double x)
{
double y = 1.0 / ( 1.0 + 0.3275911 * x);
return 1 - (((((
+ 1.061405429 * y
- 1.453152027) * y
+ 1.421413741) * y
- 0.284496736) * y
+ 0.254829592) * y)
* exp (-x * x);
}
// Returns the probability of x, given the distribution described by mu and sigma.
double pdf(double x, double mu, double sigma)
{
//Constants
static const double pi = 3.14159265;
return exp( -1 * (x - mu) * (x - mu) / (2 * sigma * sigma)) / (sigma * sqrt(2 * pi));
}
// Returns the probability of [-inf,x] of a gaussian distribution
double cdf(double x, double mu, double sigma)
{
return 0.5 * (1 + mjd::erf((x - mu) / (sigma * sqrt(2.))));
}
}
We can then call these functions within another C++ function as in the following block of code. Again, note that to actually make the function available to R, we need to include an // [[Rcpp::export]] statement on the line directly before the function definition starts.
// [[Rcpp::export]]
double Mutual_Information(
double mu,
double sigma,
double draw
){
double cdf_val = mjd::cdf(draw,mu,sigma);
return cdf_val;
}
The Boost Library
While basic C++ (pre ISO 2011) and the Armadillo libraries provide a whole lot of functionality, there are some things we may want to do -- particularly sampling from all sorts of distributions, that base C++ does not do. Fortunately, we can make use of the Boost libraries to sample from all sorts of distributions and a whole bunch of other low level stuff that can be really useful. Eventually, when R switches to using C++ (2011) by default, all of the functionality in the Boost libraries will be included in the C++ std libraries by default, but until then, we can get access to most of this functionality through the BH (short for Boost Headers) package. Once we have called the library, we can then start writing functions using the Boost features as follows. To being with, here is what my C++ file header looks like when using the boost libraries
// [[Rcpp::depends(RcppArmadillo)]]
// [[Rcpp::depends(BH)]]
#include <RcppArmadillo.h>
#include <boost/random.hpp>
#include <boost/random/uniform_real_distribution.hpp>
#include <math.h>
#include <cmath>
using namespace Rcpp;
Now lets look at some common functions we might want to grab from the boost libraries, starting with a random number generator where seed is an integer we pass in from R:
boost::mt19937 generator(seed);
We can also get a discrete distribution from boost:
boost::random::discrete_distribution<int> distribution (dist.begin(),dist.end());
int temp = distribution(generator);
We can also get a Gaussian distribution as well:
boost::normal_distribution<double> normdist(mean,var);
double my_draw = normdist(generator);
If we want a continuous uniform distribution we can get one of those as well:
boost::random::uniform_real_distribution< > uniform_distribution(0.0,1.0);
We can also take a look at the performance of native C++ (enabled by Boost) against Rcpp Sugar (essentially R functions made available to C++). We can see from the Gibbs sampler example below (Taken form Hadley Wickham's Advanced R chapter) that Native C++ is roughly twice as fast. You can access the source file here
// [[Rcpp::depends(RcppArmadillo)]]
// [[Rcpp::depends(BH)]]
#include <RcppArmadillo.h>
#include <boost/random.hpp>
#include <boost/random/uniform_real_distribution.hpp>
#include <boost/random/gamma_distribution.hpp>
#include <math.h>
#include <cmath>
using namespace Rcpp;
// [[Rcpp::export]]
NumericMatrix gibbs_cpp(int N, int thin) {
NumericMatrix mat(N, 2);
double x = 0, y = 0;
for(int i = 0; i < N; i++) {
for(int j = 0; j < thin; j++) {
x = rgamma(1, 3, 1 / (y * y + 4))[0];
y = rnorm(1, 1 / (x + 1), 1 / sqrt(2 * (x + 1)))[0];
}
mat(i, 0) = x;
mat(i, 1) = y;
}
return(mat);
}
namespace mjd{
double rgamma( double shape, double scale, boost::mt19937& rng ) {
boost::gamma_distribution<> gd( shape );
boost::variate_generator<boost::mt19937&,boost::gamma_distribution<> > var_gamma( rng, gd );
return scale*var_gamma();
}
}
// [[Rcpp::export]]
arma::mat gibbs_cpp2(int N, int thin) {
boost::random::mt19937 generator;
arma::mat mymat = arma::zeros(N, 2);
double x = 0, y = 0;
for(int i = 0; i < N; i++) {
for(int j = 0; j < thin; j++) {
x = mjd::rgamma( 3, 1/(y * y + 4),generator);
boost::normal_distribution<double> normdist(1 / (x + 1),1 / sqrt(2 * (x + 1)));
y = normdist(generator);
}
mymat(i, 0) = x;
mymat(i, 1) = y;
}
return(mymat);
}
/*** R
cat("Rcpp Sugar")
system.time({
result1 <- gibbs_cpp(1000000,10)
})
# user system elapsed
# 6.435 0.072 6.640
cat("Native C++")
system.time({
result2 <- gibbs_cpp2(1000000,10)[[1]]
})
# user system elapsed
# 3.080 0.031 3.156
*/
Common Pitfalls
There are a number of common pitfalls when working with C++ and R. Here is a certainly non-exhaustive list of them.
- The number one pitfall I have made while working with C++ and R is forgetting that while R starts vector and matrix indexes from 1, C++ (along with pretty much every other programming language), starts indexes from zero, so you need to plan accordingly. This also means if you want the last entry of a vector, you need to subtract one from the length of the vector before using the resulting number as an index.
- You will also get an error message, sometimes fatal (meaning your whole R session will crash), if you try to assign a value to a vector index that does not exist. For example if we have a vector of length 10, and try to assign a number to the 11th index, we will get a fatal error, which can be tricky to diagnose.
Another common error people make is trying to access a C++ variable outside of its scope. For example, in R, we can do this:
for(i in 1:10){ myvar <- i } cat(myvar) # prints "10" to the screenHowever, in C++, we would get an error message saying no such variable exists.
for(int i = 0; i < length; ++i){ double myvar = i; } Rcpp::Rcout << "myvar value: " << myvar << std::endl;This is because a variable assigned inside a loop (or any statement enclosed by {}) is not accessible outside of that scope. To access the variable we would need to create it outside of the loop, and then modify it in the loop, as in the following example:
double myvar = 0; for(int i = 0; i < length; ++i){ myvar = i; } Rcpp::Rcout << "myvar value: " << myvar << std::endl;Scoping can be very tricky, but fortunately the C++ compiler checks implemented by RStudio will help you diagnose these problems.
- If you want to end up distributing your C++ code as part of an R package, you will want to make sure you check out this link to help avoid a number of common errors that will trip up the C++ compilers on some of the Linux distros.
Adding C++ Code to R Packages
There are a number of very difficult to diagnose problems you can run into when including C++ code in an R package that you actually intend to publish on CRAN. Some of the most common pitfalls are documented here, in the Writing R Extensions Wiki. Most of these can be totally avoided by sticking to the data structures provided by Rcpp and using syntactic sugar, which calls R functions to do things like sampling random variables.
Some Headers Not Welcome
It turns out that not all C++ headers are portable. In particular the math.h and cmath headers should not be included in C++ code that will be part of an R package. My understanding of this is that these are very commonly used libraries, and that including one in one of your C++ files may alter the functionality of other libraries. So in general, you should remove these from your code if you want to distribute it on CRAN. In practice (distributing a package on Github, for example) I have not run in to any problems with including these headers.
#include <math.h>
#include <cmath>
Using Statements
Another issue you will need to address is ambiguity in which namespace the functions you use are coming from, if you end up using functions from the std or boost or armadillo libraries. In particular, the Writing R Extensions document suggests you use using statements when you want to include basic functions like sqrt() or exp(). Where applicable, I have added the following lines of code to my C++ files below the import statements:
using std::pow;
using std::exp;
using std::sqrt;
using std::log;
These cover most of what I need to do as it relates to machine learning, but you will just want to keep in mind which basic functions you tend to use most. This way, you can reference them by their shortened names -- like sqrt() or exp() throughout the program you are writing.
Integers, ceil(), and floor()
It is also very important to keep track of what kind of number the functions you call in C++ expect. This is most important when using the ceil() or floor() functions which can handle a float or a double, but are not explicitly defined for int's. Taking the ceil() of an integer will result in an install error on solaris and a note that you should have checked the the Writing R Extensions document from the CRAN maintainers (if you can't tell, this has happened to me). I tend to just use the ceiling() function in R where possible and then pass the result in to my C++ function (this is often useful for making sure you allocate an array of sufficient size if you are only keeping every x'th sample from a Markov chain).
Assert Statements
Asserts function much like the stop() function when you put it inside of an if() statement (to check for some bad behavior) in R, where they allow the programmer to check some condition (like that a number is greater than zero, for instance), and if that condition is not met, halt execution and return an error to alert the user that something has gone wrong. The problem is, the even if the code you write will never actually produce a situation that could lead to one of these being triggered, you still cannot put a package up on CRAN that produces one of these WARNINGs when it is built. The reason for this is that when an assert statement triggers in C++, it may lead to an uninformative error or crash R, which is not the appropriate way to handle such things. We therefore have to be careful to make sure these do not pop up. These seem to be the trickiest to deal with as you will often only get an WARNING when you run devtools::check() or R CMD check on your package on certain operating systems (typically Linux distros using the gcc compiler) or Solaris). The tricky thing is that some Linux distros will not throw this WARNING, so I would suggest using travis, which will let you test your package automatically on an Ubuntu system (using gcc) every time you commit your package to Github (which is another great reason to use Git). When this happens, you will get a WARNING that looks like this:
* checking compiled code ... WARNING
File ‘GERGM/libs/GERGM.so’:
Found ‘__assert_fail’, possibly from ‘assert’ (C)
Object: ‘Metropolis_Hastings_Sampler.o’
What this message says is that there was something tricky going on with the C++ file that was referenced (in the case above, the Metropolis_Hastings_Sampler.cpp file in an R package I was writing) which lead it to produce an __assert_ statement when it was compiled into the .o file so that it could actually be run. Unfortunately, we do not get an indication of what was causing the problem. The best way I have found to diagnose this problem is to get access to a system running the OS and compiler that caused the WARNING and then trying to build the package locally on that computer. This is essential because we need to get access to the offending .o file in order to sniff out where the problem is. Once we have tried to build the package on the offending OS using R CMD check, we can then navigate to the PackageName.Rcheck/00_pkg_src/PackageName/src directory and open up the .o file (in my case Metropolis_Hastings_Sampler.o) in a text editor and search for the appropriate assert statement in the file (in my case __assert_fail). Here is an example of the output I found in the process of debugging an R package I was recently working on:
E19__PRETTY_FUNCTION__^@_GLOBAL__I_Metropolis_Hastings_Sampler.cpp^@_ZStL8__ioinit^@_ZN4RcppL1_E^@_ZN4RcppL5RcoutE^@_ZN4RcppL5RcerrE^@_ZN4armaL9eucl_distE^@_ZN4armaL9maha_distE^@_ZN4armaL9prob_distE^@_ZN4armaL13keep_existingE^@_ZN4armaL13static_subsetE^@_ZN4armaL13static_spreadE^@_ZN4armaL13random_subsetE^@_ZN4armaL13random_spreadE^@_ZStlsISt11char_traitsIcEERSt13basic_ostreamIcT_ES5_c.clone.1^@_ZN4armaL9arma_stopIPKcEEvRKT_^@_ZN4armaL19arma_stop_bad_allocIA34_cEEvRKT_^@_ZN4armaL19arma_stop_bad_allocIA39_cEEvRKT_^@.LC12^@.LC13^@.LC14^@.LC15^@.LC16^@.LC17^@.LC18^@.LC19^@.LC20^@.LC21^@.LC2^@.LC23^@.LC5^@.LC25^@.LC26^@.LC28^@.LC31^@.LC32^@.LC33^@.LC9^@.LC30^@.LC6^@.LC34^@.LC7^@.LC8^@.LC27^@.LC29^@.LC0^@.LC1^@.LC3^@.LC4^@.LC10^@.LC11^@.LC22^@.LC24^@_ZN4Rcpp8internal16NamedPlaceHolderD2Ev^@DW.ref.__gxx_personality_v0^@_ZN4Rcpp8internal16NamedPlaceHolderD1Ev^@_ZN4Rcpp10RstreambufILb1EE8overflowEi^@_GLOBAL_OFFSET_TABLE_^@Rprintf^@_ZN4Rcpp10RstreambufILb1EE6xsputnEPKcl^@_ZN4Rcpp10RstreambufILb1EE4syncEv^@R_FlushConsole^@_ZN4Rcpp10RstreambufILb0EE4syncEv^@_ZN4Rcpp10RstreambufILb0EE8overflowEi^@REprintf^@_ZN4Rcpp10RstreambufILb0EE6xsputnEPKcl^@_ZN4Rcpp12RObject_ImplINS_15PreserveStorageEED2Ev^@R_NilValue^@R_ReleaseObject^@_ZN4Rcpp12RObject_ImplINS_15PreserveStorageEED1Ev^@_ZN4arma3MatIdED2Ev^@free^@_ZN4arma3MatIdED1Ev^@_ZN5boost6random6detail21generate_uniform_realINS0_23mersenne_twister_engineIjLm32ELm624ELm397ELm31ELj2567483615ELm11ELj4294967295ELm7ELj2636928640ELm15ELj4022730752ELm18ELj1812433253EEEdEET0_RT_S5_S5_^@__assert_fail^@_Unwind_Resume^@_ZN4Rcpp20AttributeProxyPolicyINS_12RObject_ImplINS_15PreserveStorageEEEE14AttributeProxyaSINS_9DimensionEEERS5_RKT_^@Rf_allocVector^@Rf_protect^@_ZGVZ7dataptrP7SEXPRECE3fun^@__cxa_guard_acquire^@_ZZ7dataptrP7SEXPRECE3fun^@Rf_unprotect^@Rf_setAttrib^@R_GetCCallable^@__cxa_guard_release^@__cxa_guard_abort^@__gxx_personality_v0^@_ZN3mjd3pdfEddd^@exp^@_ZN3mjd3erfEd^@_
Well this is pretty much impossible to read, but if we look closer, we can find out which line of code generate the assert statement by looking at the text directly before the __assert_fail statement. I this case we have:
@_ZN5boost6random6detail21generate_uniform_realINS0_23mersenne_twister_engineIjLm32ELm624ELm397ELm31ELj2567483615ELm11ELj4294967295ELm7ELj2636928640ELm15ELj4022730752ELm18ELj1812433253EEEdEET0_RT_S5_S5_^@__assert_fail
which upon further inspection originated in the generate_uniform_real function. By going and looking up the header file for this function on the internet, we can see that there are a number of assert statements in the source code. From here we basically have two options:
We can try to remove the dependency of our code on this function. This is often the easiest solution and probably the best, as we can just search for other alternatives that do not need to do so much checking. In the example I have been using, I ended up replacing
boost::random::uniform_real_distribution; uniform_distribution(0.0,1.0); which required two endpoints for the distribution that had to be checked to one that did not require any checking, which thus removed the assert statement:
boost::uniform_01uniform_distribution; solving my problem. This approach is very case specific but can often be a simple and fast workaround.
You can also attempt to rewrite the offending function manually to remove the assert calls from the source code. This is more risky as you now have to do all of the checking yourself, but can also be very rewarding as you get to use exactly the function you wanted to. I ended up doing this for the boost
normal_distributionfunction which contains the following line of code:BOOST_ASSERT(_sigma >= RealType(0));which was causing gcc to throw a WARNING which R picked up and would not let my package pass
R CMD check. I simply removed the offending line of code and then included the header file in my package but under a different namespace (replacingnamespace boost {withnamespace mjd {in my case) so that the function could now be called using:mjd::normal_distributionnormdist(mean,var); which will give identical performance to the boost version but not throw a WARNING. This just means we need to ensure that we never pass a non-positive variance to the normal distribution, so a check should be done in R or in C++ to make sure this does not happen.